7. Training models with DeepProfiler#
7.1 Export single cells#
Imagine you already have a dataset of full images and you would like to train a model. Models are trained with single-cell crops, so the single cells of the dataset should be exported separately. If the dataset consists of 16-bit TIFF images, it is strongly recommended to pre-process the dataset first.
The single-cell export tool requires single cells to be identified and segmented ahead of time (see Section 3). Then, this tool will generate an image for each existing single-cell in the locations file with the channels concatenated in horizontal order (see example image below). The basic command for export is as follows:
python deepprofiler --root=/home/ubuntu/project/ --config=export.json --metadata=index.csv --single-cells=single_cells_dataset export-sc
Where --root is the root directory of your project, --config is the name of the configuration file that you want to
use for this command, --metadata is the name of the metadata file listing the images you want to process with this
command (note that it can be a subsample), and --single-cells is the name of the directory that will be created for
storing the output.
Fig. 5 Console output when export finishes.#
The exported images and metadata (sc-metadata.csv) will be stored in /project/outputs/single_cells_dataset/
(you control the name of this directory with the flags described above). If the single-cells parameter is not set, the
default folder name for the exported dataset will be single-cells. The size of the cropped region is defined by a box_size
parameter in the configuration file. The images are saved as a stripe of crops from each channel (as listed in the channels
field in the configuration). Optionally, cell masks can be extracted, it would appear last in the stripe.
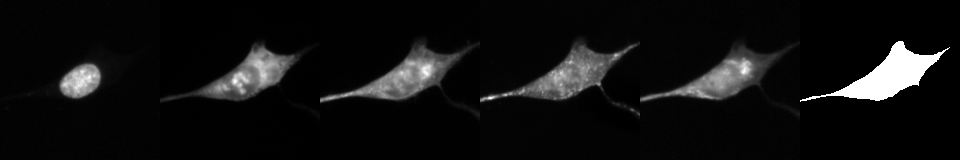
Fig. 6 Example of exported image from BBBC037 dataset.#
About training and validation splits
As mentioned in the config description, the metadata field split_field and its values from training and validation configuration config fields define the initial training-validation split, which will be reflected in the single-cell metadata file for each record.
If a different training-validation split is needed, you can modify the split_field column (for example with pandas) and save the new metadata to a different file and then use it for training, there is no need to export images again. It is also possible to append a new column for a new training-validation split to an existing metadata file, in that case, don’t forget to change split_field in the training configuration file.
7.2 Train a model:#
An example of the training command:
python3 deepprofiler --root=/home/ubuntu/project/ --config filename.json --single-cells=single_cells_dataset --exp=experiment_name --gpu 0 train
Training arguments
The default single-cell metadata file is sc-metadata.csv, though it can be different by passing --metadata parameter.
--gpu parameter is an id of a GPU to be used, available GPUs with their IDs can be listed with nvidia-smi command.
The --config filename.json parameter points to the name of a file from /inputs/config folder.
The --exp experiment_name points to a folder in /outputs/ folder.
In the beginning the ImageNet pre-trained weights (if initialization is set to ImageNet in the config)
are assigned and you will see the following console output:

Fig. 7 ImageNet pre-trained weights are beings assigned. It can take a little time.#
Then training will start.
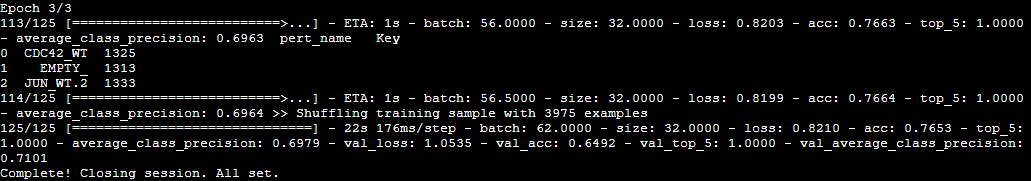
Fig. 8 Example of the training output with a closing message.#
Training checkpoints will be saved in /project/outputs/experiment_name/checkpoint/, the logs with accuracy and losses
in /project/outputs/experiment_name/logs/.
If you run example data training, you would get approximately reproduce the following classification results:
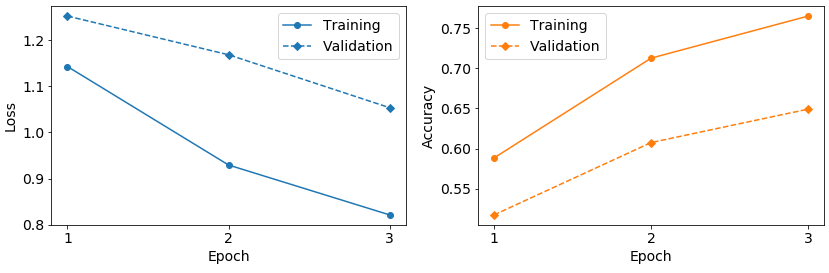
Fig. 9 Loss and accuracy over epochs while training on example data.#
Crop generators
Crop generator plugins define the way how the data is going to pass through models. The default choice in most cases is sampled_crop_generator. You can explore other available crop generators or create your own.
About class balancing
DeepProfiler performs class balancing, so no need to worry about an initial class imbalance in the processed dataset.
