4. Metadata and single-cell locations#
DeepProfiler requires metadata files that provide information about the experiment as well as the locations of single cells
(centroids) in your images. These files are stored in project/inputs/ and there are two main types:
A metadata file (located in
project/inputs/metadata/) that relates image files to the experiment structure (e.g., plate and well positions) and includes additional information about replicates and treatments. The default filename of the metadata file isindex.csvif the--metadataparameter was not explicitly set.--metadatais the metadata’s filename inproject/inputs/metadata/.A series of single-cell locations files for each site (located in
project/inputs/locations/) that provide the XY coordinates of nuclei centroids. Note that this file is not required if running DeepProfiler infull_imagemode.
4.1 The metadata file#
The metadata file is critical for running DeepProfiler. It follows a comma-separated-values (CSV) format with a header, contains information about the experiment, and lists all images in your project. DeepProfiler uses this file to guide image sampling for running learning algorithms and to find the images that we want to process. This file is expected to contain metadata to identify the context of images in the physical experiment that produced them, for instance, identifiers of plates, wells and fields of view (i.e., sites; see Fig. 3).
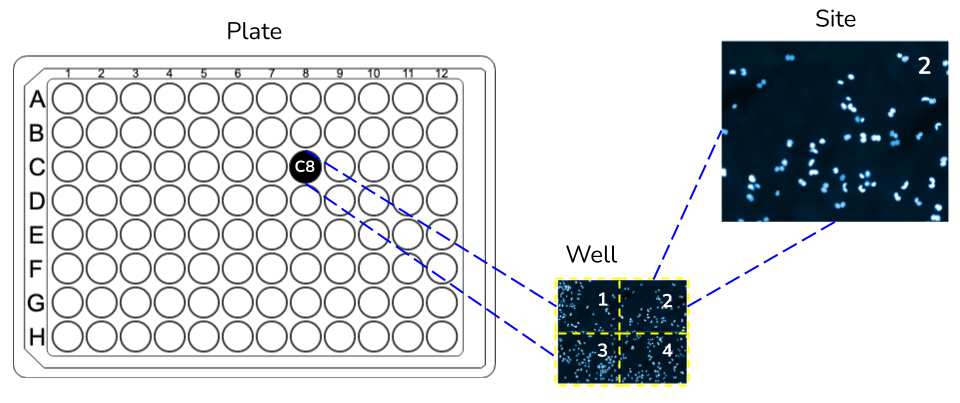
Fig. 3 Schematic of plates, wells and sites, which are three metadata fields required by DeepProfiler in the metadata file.#
DeepProfiler assumes that each row in the file represents one (multi-channel) field of view. The following list indicates the minimal columns that the metadata file is expected to have:
Metadata_Plate: Name or identifier of the plate (i.e., the highest level of experimental organization), e.g.41744. The field header cannot be renamed.Metadata_Well: Position in the plate, e.g.A21(i.e., the middle level of organization within plates). The field header cannot be renamed.Metadata_Site: A microscope acquires images in different sites within each well (i.e., the lowest level organization within wells). For instance, sites may cover a 4x4 grid or a 9x9 grid, depending on resolution and other factors. The site identifier for each image goes here, e.g.3. The field header cannot be renamed.Channel_Name: Relative path to the image file of each channel. An experiment may have multiple imaging channels (i.e., colors) and DeepProfiler assumes that each channel is stored in a separate image file (all channels stored in the same file are not currently supported). Therefore, to put together all the channels of a single image, the metadata file will need to have multiple channel_name columns listed in the configuration file. These columns can be renamed as necessary and should point to the corresponding image files using a path relative to the image directory. For example, an assay with DNA, RNA, and Mito stains will have three channel columns named accordingly, with entries in each column pointing to the corresponding image file. The field may have different names.Image path (default): The images can be located in the default DeepProfiler folder (which is
inputs/images) so no specific path needs to be defined here, only the Plate/FileName, as shown in the metadata file example.Image path (customized): If the user cannot move the images to the default location, images can also be located in a specific folder inside the user’s machine instead of the default. For example, a user can specify the location of the DNA image to be
C:\Users\User1\images\41744\taoe005-u2os-72h-cp-a-au00044859_f21_s1_w159ff0023-3fa4-4acb-a277-af596b4e9e25.tif.
Treatment: We assume that cells in a well have been treated in a biologically meaningful way or represent different experimental conditions. This column keeps track of that information, which may have other names (in the provided example data, it is calledpert_name). It is useful as an identifier of the type of biological experiment, treatment, perturbation or condition of cells observed in the images. This column must be a biologically meaningful label that could be used by DeepProfiler for training purposes. May have different names.Replicate: Number or identifier indicating which repetition of the treatment an image corresponds to. In the provided example data this column is calledpert_name_replicate.
These are the minimum columns required in the metadata file. You can append more columns with information specific to your experiment as needed, to keep track of other metadata in your project. We recommend that if you do add additional columns, their names should not include any spaces. Notice that the order of columns is not important, as long as these are available. The meaning of the columns can be interpreted differently according to your problem, for instance, instead of plates, you may be interested in subjects or patients. However, the three levels of an organization (plate, well, site) are expected, even if you don’t explicitly use them (e.g. set wells to a constant string if it does not apply to your data). The name of certain columns can be changed as well and later associated with the expected information in the configuration file (Section 5).
Click here to see an example metadata file (See also Fig. 4)
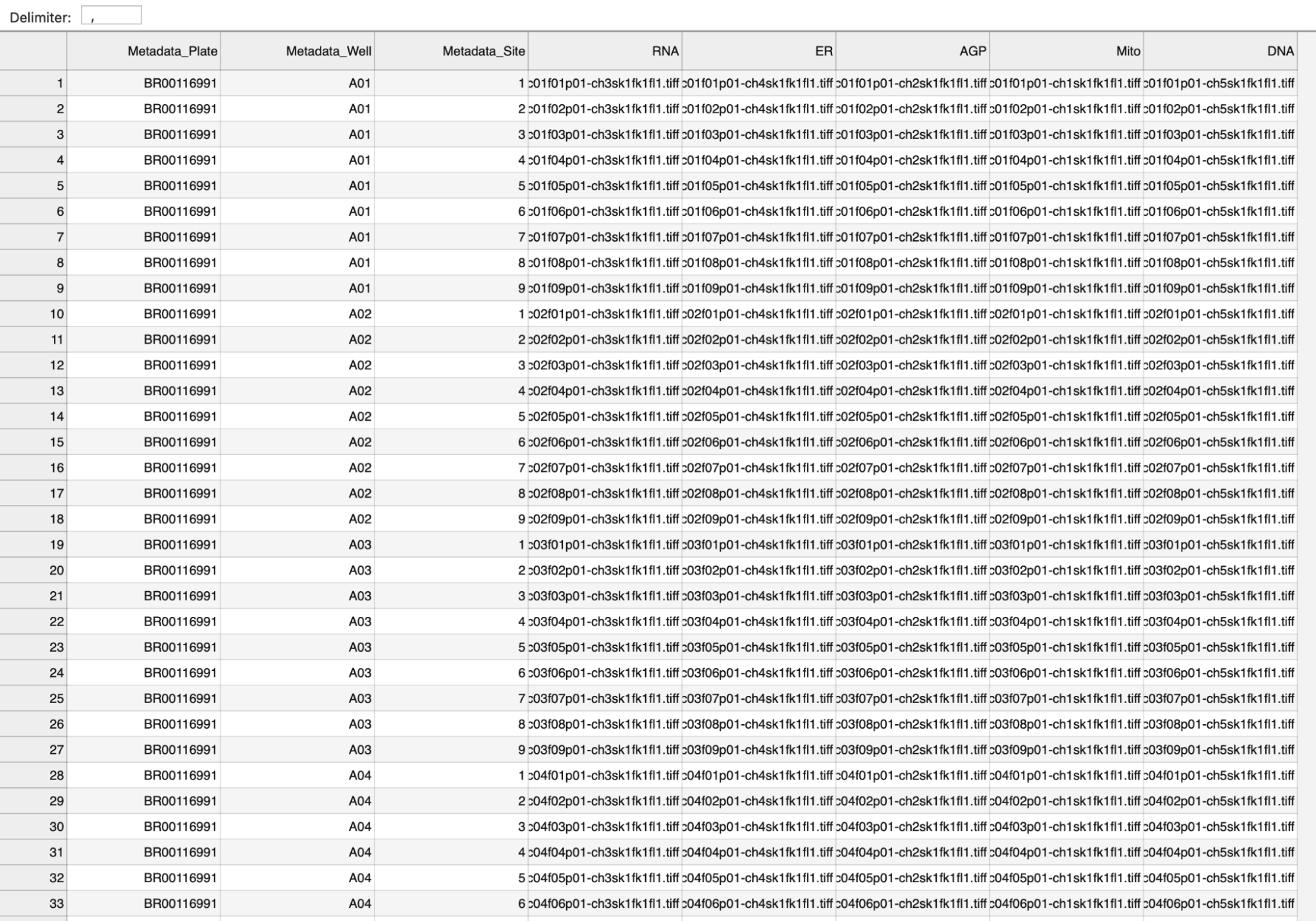
Fig. 4 Screenshot of an example metadata file#
4.2 Single-cell locations file#
DeepProfiler needs the x and y coordinates for the centers of nuclei in your images. There should be one location’s file
for each site (i.e., each image) in your experiment. These files need to be stored in a particular structure:
project/inputs/locations/{Metadata_Plate}/{Metadata_Well}-{Metadata-Site}-Nuclei.csv. The Metadata_Plate, Metadata_Well
and Metadata_Site must match what is in the metadata you currently use from project/inputs/metadata/.
Here is an example of how to structure the location’s data:
.
└── project/
└── inputs/
└── locations/
└── Plate1/
├── A01-f01-Nuclei.csv
└── A01-f02-Nuclei.csv
...
The contents of each location’s CSV file for a particular site should be just two columns headed with Nuclei_Location_Center_X
and Nuclei_Location_Center_Y. The pixel coordinates of centers for each nucleus should be listed in these columns (see below):
Nuclei_Location_Center_X,Nuclei_Location_Center_Y
219,34
175,45
1065,51
These coordinates can be generated by a program like CellProfiler if you segment nuclei with a module like IdentifyPrimaryObjects.
